Research
 My primary research interests are in evolutionary microbiology. I have a strong background in evolutionary bioinformatics and have examined problems related to the origin and genome evolution of eukaryotic microbes (a.k.a. protists) and archaea.
My primary research interests are in evolutionary microbiology. I have a strong background in evolutionary bioinformatics and have examined problems related to the origin and genome evolution of eukaryotic microbes (a.k.a. protists) and archaea.
To address those questions, I mainly use phylogenetics/phylogenomics and comparative genomics approaches.
Research overview
During my career, I have acquired expertise in phylogenetic placement of novel lineages and investigation of deep phylogenetic relationships, as well as in reconstructing the evolutionary history of key cellular systems at the scale of eukaryotic diversity.
I have also applied Bayesian relaxed molecular-clock methods to estimate the age of the different eukaryotic supergroups, and to time the origins of multicellularity.
In the last few years, I have become increasingly interested in the role of horizontal gene transfer (HGT) in eukaryotic genome evolution.
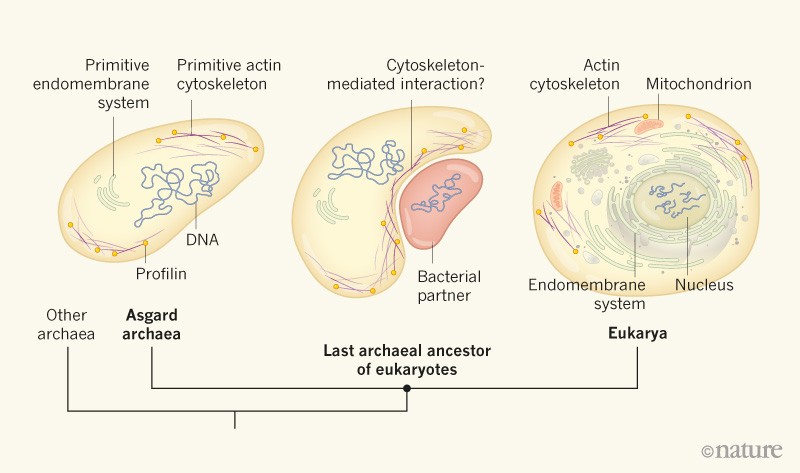
Finally, I have long been interested in archaeal evolution. I have worked in the field of archaeal evolutionary genomics, specifically focusing on the newly discovered Asgard superphylum, from which eukaryotes are thought to have evolved.
Past and current projects
March 2019 – to date Principal Investigator (chargée de recherche, CNRS)
Diversity, Ecology and Evolution of Microbes group (Ecology, systematics and evolution Unit). Université Paris-Sud.



My research focuses on the evolution and diversity of epigenetics, and on horizontal gene transfers in eukaryotic genomes, with a emphasis on microbial eukaryotes (protists). For this project, we use (single-cell) genomics, phylogenetics and comparative genomics approaches. This project is funded by an ERC Starting grant (Macro-EpiK, 03/2019-02/2024).
We are looking for Masters, PhD students and postdocs. Don’t hesitate to send me an email if you are interested.
Jan 2017 – Feb 2019 Marie Curie Fellow
Molecular Evolution Group (Dpt of Cell and Molecular Biology Uppsala University, Sweden). Supervisor: Dr. Thijs Ettema
Prokaryotic origins of eukaryotes
At the cellular level, the gap between prokaryotes and eukaryotes is immense, with the latter cell types displaying a large number of complex subcellular organelles and molecular systems. Although many eukaryotic-signature proteins (ESPs) are known to trace back to the last eukaryotic common ancestor, their deeper evolutionary origins remain unclear. My major goal is to illuminate the prokaryote to eukaryote transition by elucidating the origin and subsequent evolution of ESPs, as well as their order of emergence. In addition, I aim to better pinpoint the placement the eukaryote lineage within the tree of Life using state-of-the-art phylogenomic approaches.
June 2011 – Dec 2016 Post-Doctoral Fellow
Centre for Comparative Genomics and Evolutionary Bioinformatics (Halifax, Canada). Supervisor: Dr. Andrew J. Roger.
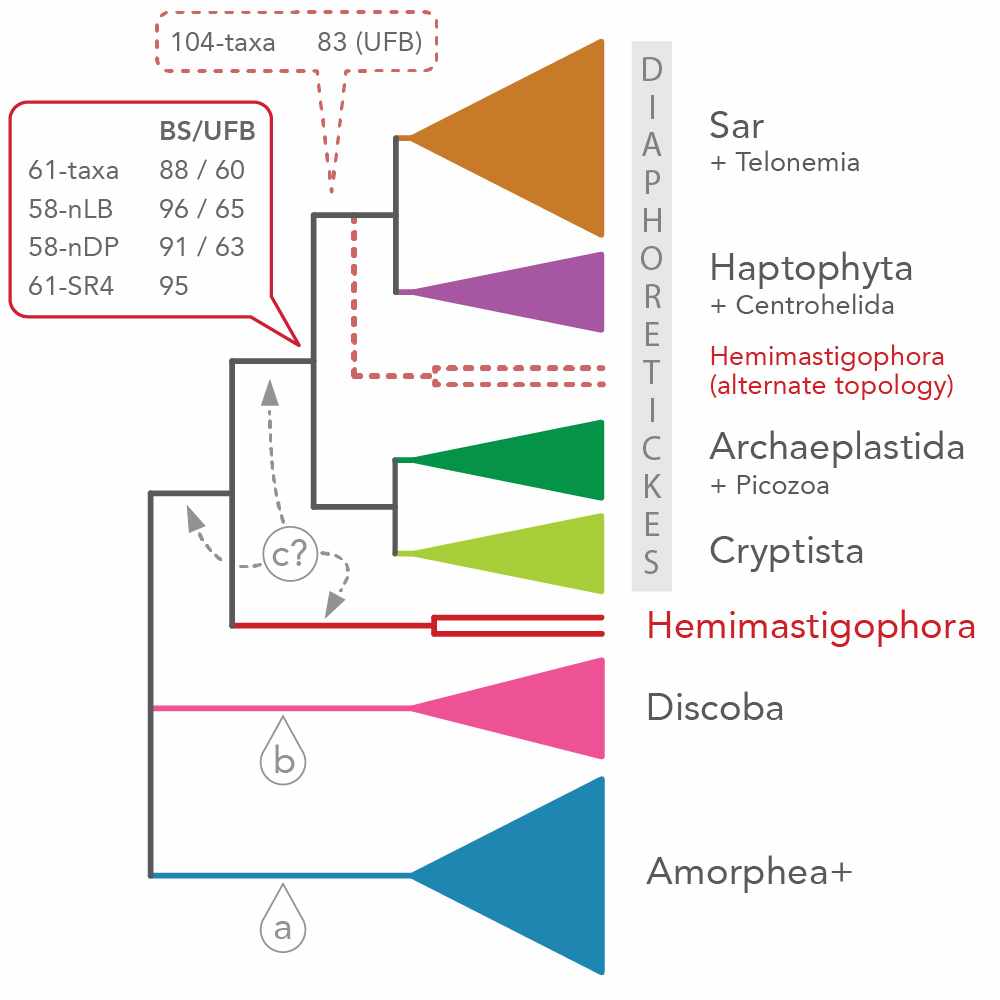
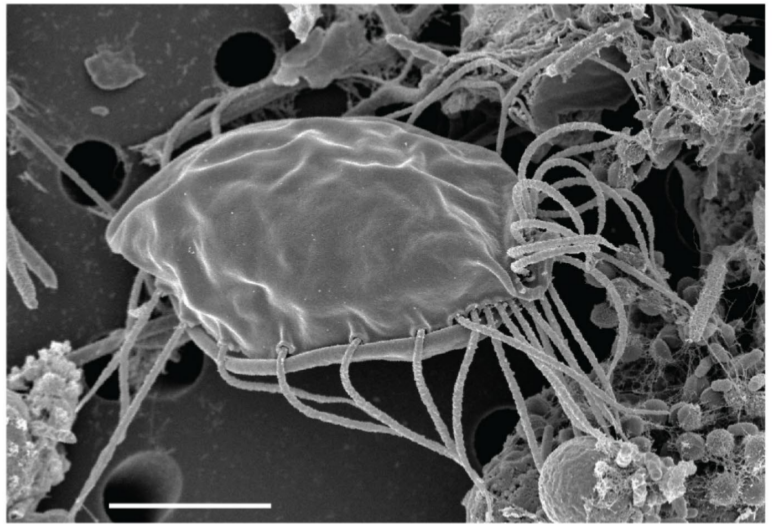
Root of the Eukaryotic Tree
One of my major interests lies in the investigation of the relationships amongst eukaryotic lineages as well as the position of the root of the eukaryote tree. Eukaryotes are usually divided in six ‘super-groups’, but this hypothesis is still controversial. One of my postdoctoral projects aims to revisit the problem of robustly rooting the tree of eukaryotes through the investigation of three kinds of untapped phylogenetic markers.
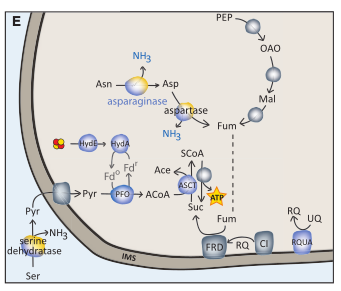
Adaptation of eukaryotic microbes to anaerobiosis
I have been interested in the evolutionary transition from aerobic to microaerobic/anaerobic conditions that occurred several times independently across the tree of eukaryotes. In particular, I am interested in how this transition has affected mitochondrial functions and has reshaped the proteomes and biochemical abilities of the mitochondrial-related organelles (MROs) found in anaerobic protists.
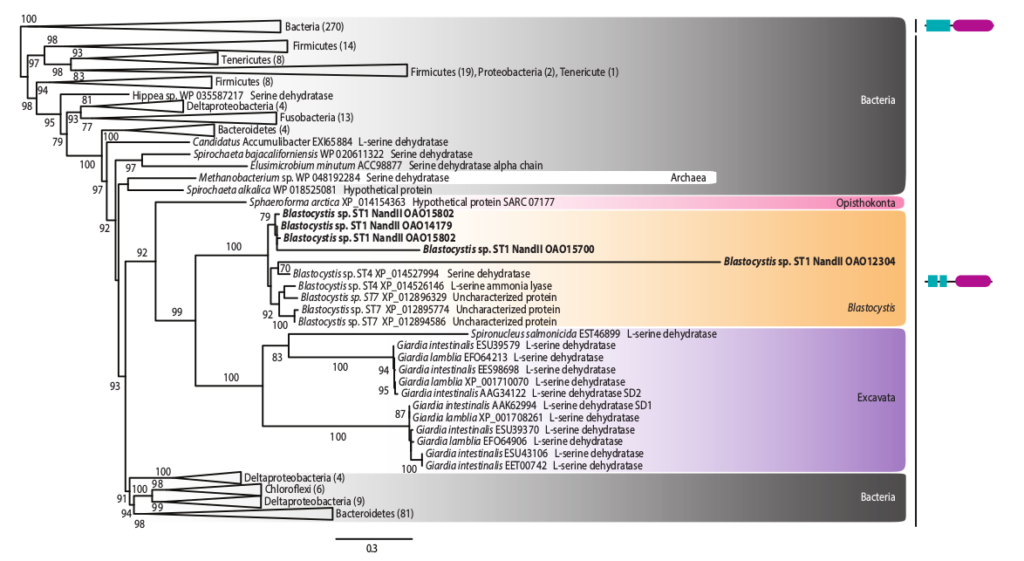
Evolution of parasitic protists
I am also interested in the genomic adaptations occuring during the transition from free-living to parasitic lifestyles, and in particular the ones in relation with the acquisition (through lateral gene transfers) and expansion of genes involved in anaerobiosis, pathogenicity, nutrient acquisition and response to host defense mechanisms.
Eukaryotic microbes in the human microbiome
Recently, I have been interested in the impact of eukaryotic microbes (parasites and commensals) on the human gut microbiome. Since the prevalence and roles of microbial eukaryotes has been rarely studied, we aim to obtain a better understanding of the relationships between particular parasitic infections and pathologies such as inflammatory bowel disease (IBD).
October 2007 – June 2011 PhD in Phylogeny, Evolution and Bioinformatics.
Team Genome, Evolution and Bioinformatics (Laboratoire de Chimie Bactérienne, CNRS, Marseilles, France). Supervisor: Dr. Céline Brochier-Armanet.
Characterization of LECA — Evolution of eukaryotic multiprotein complexes
The development of high throughput techniques, especially in proteomics and genomics has yielded extensive data that can be used in evolutionary analyses. In this context, eukaryotic multiprotein structures (EMS) are objects of interest. Indeed, these large protein complexes are involved in many fundamental processes of eukaryotic cells, and have no homologues in prokaryotes (even if the functions in which they are involved may exist) and therefore have certainly played a major role in the eukaryogenesis.
During my PhD, I carried out the phylogenomic analysis of two EMS involved in cell division (the midbody and the APC/C) and showed that these systems have an ancient evolutionary origin and were already present in the last common ancestor of eukaryotes (LECA), while resulting from eukaryotic innovations. This implies that the emergence of these two EMS occurred after the divergence of the eukaryotic lineage and before the diversification that gave rise to the current lineages.
Beyond these evolutionary questions, analyses of these EMS uncover some biological aspects of these systems. Indeed, if these systems were generally well conserved during the diversification of eukaryotes, their analysis shows a high plasticity of composition in some protist lineages. This suggests recent changes regarding certain phases of these organisms cell cycle which would be interesting to explore experimentally.
Eukaryotic tree of life
Concomitantly, this work showed that, although being operational protein, components of these EMS carry a phylogenetic signal usable for inferring phylogenetic relationships among eukaryotic lineages. Construction of supermatrixes from these proteins led to the inference of phylogenies of high quality. Combining these data with those from analyses based on informational proteins showed a significant progress on the resolution of inferred trees. These results open the field of possibilities to find other markers among the untapped operational proteins.
January – June 2007 Master degree internship in Bioinformatics and Evolution.
Development of a methodological approach for the study of EMS. Team Genome, Evolution and Bioinformatics (LCB, CNRS, Marseilles, France). Supervisor: Dr. Céline Brochier-Armanet.

Comments on "Research"