News
March 2024

🧂 Our latest study unravels the complex evolutionary trajectories of extremely halophilic archaea, organisms that thrive in near-saturation salt concentrations.
🌊 Exploring metagenomes from the hypersaline aquatic systems of the Danakil Depression, Ethiopia, we’ve discovered two previously unknown lineages of extreme halophiles, Afararchaeaceae and Asbonarchaeaceae, for which we reconstructed MAGs and predicted basic metabolic features
🖥 Using state-of-the-art phylogenetic models and approaches to overcome the amino acid composition biases inherent to adaptation to high salt environments, our analysis suggests at least four independent evolutionary adaptations to extreme halophily in Archaea
🔄 Finally, using reconciliation approaches, we show that gene duplication and horizontal gene transfer appear pivotal in these adaptations, spreading key genes across these lineages

Danakil Depression, Afar region of Ethiopia. Credit: Eric Lafforgue
January 2024
Jolien Van Hooff submitted the second paper of her postdoc work on the origin and evolution of eukaryotic SMC complexes.
Structural Maintenance of Chromosomes (SMC) complexes play integral roles in maintaining the integrity of the genome and the proper functioning of cells. Operating in chromosome organization and dynamics, they ensure the proper segregation of chromosomes during cell division and participate in processes like DNA damage repair, replication, and gene regulation.
Although SMC complexes are fundamental to cellular biology, research revealed significant variations in their functions and compositions across different organisms. This variation in SMC complex composition and roles raises intriguing questions about the true diversity and evolution of SMC complexes across eukaryotes, and their deeper, prokaryotic evolutionary origins. Indeed, while most prokaryotes possess only one SMC complex, eukaryotes usually have four. Understanding the origin of such crucial components of eukaryotic cells is deeply linked to understanding the establishment of the eukaryotic cell itself, including the nucleus and linear eukaryotic chromosomes.
In this study, we conducted extensive comparative genomic and evolutionary analyses of SMC complexes across various eukaryotic and prokaryotic species. Our findings can be summarized around three main points:
- One of the SMC complexes, condensin II, was lost frequently throughout the eukaryotic tree, and these losses may underpin the observed variability in nuclear genome architecture among different lineages.
- The last eukaryotic common ancestor (LECA) already possessed four distinct SMC complexes, with virtually identical compositions as the complexes found in modern-day eukaryotes.
- LECA’s SMC complexes evolved during eukaryogenesis from two distinct complexes inherited from its Asgards archaeal ancestors. Hence, these archaea already possessed more than one SMC complex, in contrast to many other prokaryotes. This finding bolsters the hypothesis that many intricate cellular characteristics of eukaryotes evolved from simpler building blocks inherited from their Asgard archaeal ancestors.
December 2023
Jazmin Blaz beautifully defended her Ph.D. today! It’s been very special to watch her mature and take ownership of her (very ambitious) project. Our team is grateful that she decided to take a leap of faith and cross the Atlantic for this adventure. Congratulations, Dr. Blaz!

September 2023
Dr. Eme defended a very French degree, the “authorization to direct research” (Habilitation à Diriger la Recherche). It is the highest degree awarded in France, and “recognizes the high scientific level of the candidate, the original character of their approach in a field of science, their ability to master a research strategy in a sufficiently broad scientific field and their ability to supervise young researchers.” (Let’s put aside the irony of having supervised students to obtain the degree that allows you to supervise students). It was an opportunity to have a stimulating discussion about the origin and evolution of eukaryotes with a fantastic panel compose of Drs. Alastair Simpson, Toni Gabaldon, Jaime Huerta-Cepas, Céline Brochier-Armanet and Olivier Lespinet.

June 2023
Laura and her collaborators just published the results of a 5-year effort undertaken in Thijs Ettema’s group on the Asgard archaeal ancestors of eukaryotes, combining metagenomics, phylogenomics, ancestral reconstruction, and metabolic inferences.

A thread:
February 2023
The lab is leaving for the second time for a field trip in Chile, this time in the Southern part of the Atacama region. We aim to sample the microbial diversity of unusual environments such as high-salt ones.

September 2022
PhD candidate Kelsey Williamson (Dalhousie University, Canada) is visiting the lab for a few weeks as part of a collaboration between Andrew Roger’s lab and the Eme group on deep eukaryotic phylogeny funded by the Simons Foundation. Welcome, Kelsey!

September 2022
Laura, Jazmin, Jolien and Fabian participated in the EMBO Comparative Genomics of Unicellular eukaryotes workshop in San Feliu, Spain.
Jolien van Hooff and Jazmin Blaz were selected for presenting posters on their current work on Lateral Gene Transfer in Rhizaria, and comparative genomics of ancyromonads, respectively.
Fabian van Beveren was selected for an oral presentation on the origin of red complex plastids.
Laura Eme was an invited speaker and talked about the genome of a deep-branching eukaryote belonging to the CRuMs, Mantamonas.
May 2022
Jazmin participated in the Evo-chromo EMBO workshop in Aarhus, Denmark, where she got to present her PhD work on the (epi)genome of ancyromonads.

April 2022
The first paper out of Fabian van Beveren’s Ph.D. work is out! We sequenced plastid and mitochondrial genomes from a large number of unrepresented red algal lineages and explore the reasons for the dramatic size differences. Read the full story here open access in GBE!

December 2021
Jolien van Hooff was awarded the prestigious Veni Fellowship. Congratulations, Jolien, very well deserved!


March 2021
New paper in collaboration with the Waller Lab! Happy to have been a small part of this adventure. Read the full story here in PLoS Biology! A thread by Ross:
Dec 14, 2020
Laura was elected at the Secretary position of the International Society of Evolutionary Protistology (ISEP) for a 2 year term.

August 3, 2020
We’re hiring a postdoc in phylogenetics! More specifically to work on investigating methodological artefacts affecting inferences of deep eukaryotic evolution.
The position is in the DEEM Team at the University Paris-Saclay, and in close collaboration with Andrew Roger and Ed Susko (Dalhousie University, Halifax, Canada), and Minh Bui (ANU, Canberra, Australia).
Aug 1, 2020
We’re hiring!

6 month Master’s student project – Lateral gene transfers between closely related eukaryotes
Background
Horizontal gene transfer, also called lateral gene transfer or LGT, allows the acquisition of genetic material, and thus new functions, from distantly related organisms. This process is well known to be a major driver of evolution in prokaryotes, and is for example responsible for the acquisition of antibiotic resistance in bacteria. In contrast, the role and frequency of LGT in eukaryotes has been much less studied, although there are clear examples of its impact on the evolution of certain unicellular eukaryote parasites. When they exist, these recent studies often focus on gene transfer events from very distant organisms, such as from bacteria, because they are easier to detect with confidence. However, it has been shown in bacteria that LGTs more often occur among closer related lineages. This can be explained in part because horizontally acquired genes that use codons that better match the codon usage of their new host genome will be better optimized for expression within the context of that genome and will confer less of a fitness cost on their new host. . In eukaryotes, the extent to which closely related organisms exchange genetic material and how much this varies across the diversity of eukaryotic lineages is, in contrast, limited.
Project
This project aims to estimate the importance of LGT between closely related eukaryotic species. It consists of two parts: A) developing an approach that detects genes that are very likely to be derived laterally from a different, but closely related eukaryote, and B) using this approach to study which eukaryotic clades underwent many of such ‘short-distance’ LGTs andto identify the features (such as function or codon usage) of genes shared via LGT between closely related species. The developed approach will first be benchmarked with a set of published LGTs, and will then be applied to identify cases of LGTs in understudied lineages of unicellular eukaryotes (protists).
Your profile
Ideally, the candidate will have some knowledge in phylogenetics/molecular evolution and some knowledge or strong desire to learn scripting (Python or Perl) and working in a linux environment. Additional requirement: good level in English, good oral and written communication skills, autonomy, organization skills.
Practicalities
This is a 6 month internship that will take place between January and June 2021. A financial compensation of €577.50/mo is offered.
The research unit of “Ecology, Systematics and Evolution” (ESE) is located in the University Paris-Sud campus in Orsay. This campus is 20 km south of Paris and well connected through commuter train.
The student will work within the DEEM (Diversity, Ecology and Evolution of Microbes: http://www.deemteam.fr/en) team, which has a broad expertise in ecology and evolutionary biology, with a focus on microbes (prokaryotic and eukaryotic). This internship will be co-supervised by Dr. Laura Eme (laura.eme@universite-paris-saclay.fr), principal investigator, and Dr. Jolien van Hooff, postdoctoral fellow (jolien.van-hooff@universite-paris-saclay.fr).
Note the possibility to apply for a PhD scholarship from the doctoral school at the end of the internship if you are interested in pursuing a PhD with us.
Interested?
If you are interested in genome evolution and pursuing a research project in bioinformatics and phylogenetics, please contact Dr Laura Eme (laura.eme@universite-paris-saclay.fr) for more information or apply directly by sending your CV and a cover letter explaining your motivation for undertaking this project.
June 27, 2019
Update: this position is now filled. Get in touch if you want to apply for a scholarship/fellowship with me.
PhD position available: (epi)genomics of protists
Practical information
Location: Paris-Sud University (Orsay, France)
Type of position : PhD position (full time)
Contract length : 3 years (possibility of a one year extension)
Starting date : Oct 1st, 2019 (negotiable)
Annual gross salary: € 25,620
Application deadline : July 17, 2019
Project overview
The candidate will work in the context of the ERC Starting Grant Macro-EpiK, which aims, among others, at better understanding the diversity and evolution of epigenetic modifications in eukaryotic microbes (protists).
During his/her PhD, the candidate will take part in 1) preparing genetic material (RNA and DNA) for transcriptome, genome and methylome sequencing, and 2) their subsequent assembly, annotation and various other bioinformatic investigations.
More specifically, they will be in charge of the following tasks:
– preparation and maintenance of protist cultures, with an emphasis on under-studied lineages;
– RNA/DNA extraction for various NGS sequencing techniques (Illumina, Nanopore, Hi-C, bisulfite…), from cultures or, in the case of organisms difficult to grow at high density, directly from single-cells;
– sequence assembly and annotation in order to obtain high-quality genome data;
– bioinformatic investigation of the epigenome;
– phylogenetic reconstruction of specific genes of interest.
Your profile
We will prioritize candidates with an excellent background in bioinformatics (transcriptome and/or genome analyses including assembly, data cleaning, annotation, and molecular phylogenetics). Alternatively, we will consider candidates with microbiology background (culture maintenance, genetic material preparation for sequencing), under the condition that they show a real enthusiasm to acquire an expertise in bioinformatics. This is an ambitious project which will require commitment and thoroughness.
Additional requirements
– English proficiency (written and spoken).
– Good presentation and communication skills
– Autonomy and organization
– Synthetic mind and critical thinking
Working environment
The Ecology, Systematics and Evolution (ESE) unit is coaffiliated CNRS – Paris-Sud University – AgroParisTech, and is located on the Paris-Sud campus, in Orsay. Orsay is 20 kms South of Paris and directly accessible by the RER (commuter rail).
The candidate will be a member of the DEEM team (Diversity, Ecology et Evolution of Microbes: http://www.deemteam.fr/en), which focuses on microorganismal diversity and evolution, using molecular phylogenetics and comparative genomics. The candidate will work closely with other members involved in the Macro-EpiK project and will be supervised by Laura Eme (laura.eme@u-psud.fr).
Our team is international and the working language is English. We work in a good atmosphere and an inclusive and supportive environment. We welcome candidates from all horizons!
Applications
!! Please apply using the following link : http://bit.ly/2KGrcDU!!
We will not be able to consider direct applications by email. Feel free to contact me (laura.eme@u-psud.fr) for any additional information. Applications should include a detailed CV, a cover letter and contact information for at least two referees.
May 2019
In two studies recently published in Nature Microbiology and Nature Communications, we have investigated the potential metabolic abilities of Asgard archaea and explored the theoretical implications for the origin of eukaryotes.
March 28, 2019
The ceremony for the medical research prizes of the Fondation de France took place last night. I am particularly thankful to the Fondation Jacques Monod for its engagement toward basic research and for its decision to award its prize for the first time to the field of evolutionary biology.
All laureates and a summary of the evening is available here (in French):

A talented scientific illustrator summarized some of my research on eukaryotic origins (©Fondation de France / Sophia Lemonnier):


Photos: © Fondation de France / Anthony Guerra
December 20, 2018
Honoured to be awarded the 2018 Jacques Monod Prize!
The Jacques Monod Foundation is under the authority of the “Fondation de France” and awards its annual prize to one or two young researchers working on molecular aspects of cell regulation.
A ceremony will be held at the College de France on March 28, 2019 for the 50th anniversary of the Fondation de France.

November 14, 2018
Using culture-independent transcriptomics and phylogenomics, our latest work published in Nature shows that the understudied and beautiful hemimastigophoreans do not belong to any known supergroup of eukaryotes.

October 3, 2018
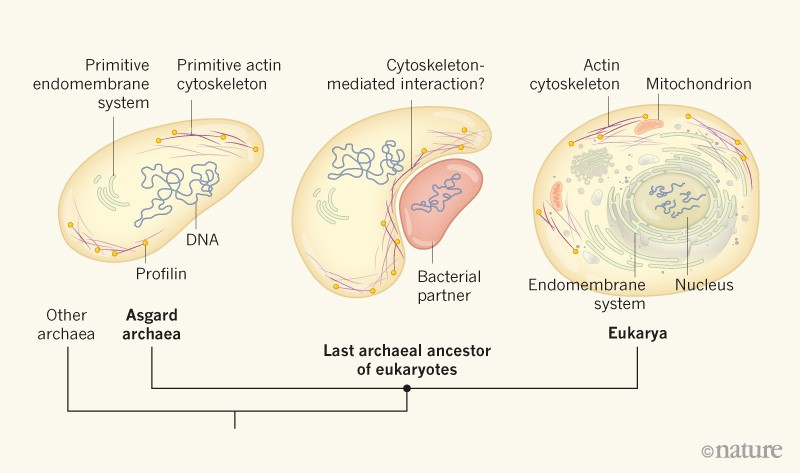
Thijs Ettema and myself have co-authored a News and Views highlighting the publication by Akil and Robinson in the last issue of Nature. In this work, the authors performed in vitro experiment on several Asgard profilins and showed that they were able to regulate the polymerization of eukaryotic actin filaments. This suggests that Asgard archaea also have a (perhaps primitive) dynamic profilin-regulated actin cytoskeleton.
July 27, 2018

Thrilled to announce that I am the recipient of an ERC Starting Grant (Macro-EpiK). I will start my research group at Paris-Sud University early 2019 as part of the Ecology, Systematics and Evolution department.
My research will focus on the evolution and diversity of epigenetics, and on horizontal gene transfers in eukaryotic genomes, with a emphasis on protists.
July 19-29, 2018
Joining the MBL Molecular Evolution workshop faculty members for the first time this year in beautiful Woods Hole, Massachusetts.

June 04, 2018
Archaeal diversity and evolution featured in The Scientist with interviews from many researchers in the field

June 03, 2018
The 2018 meeting of the International Society of Evolutionary Protistology has been a huge success. Inspiring talks from promising young researchers and world-class leaders in beautiful Cyprus. Looking forward to the next edition!
May 02, 208
Happy to announce that I have recently become an Associate Editor for BMC Genomics (“eukaryote microbial genomics” section) and Research in Microbiology.
Apr 26, 2018
Our paper on the remodelling of mitochondria by lateral gene transfer during adaptation to hypoxia in microbial eukaryotes is now out in eLife. Featuring outstanding work from Courtney Stairs.
Stairs, C.W., Eme, L., Muñoz-Gómez, S., Cohen, A., Dellaire, G., Shepherd, J.N., Fawcett, J.P. and Roger, A.J., 2018. Microbial eukaryotes have adapted to hypoxia by horizontal acquisitions of a gene involved in rhodoquinone biosynthesis. eLife, 7, p.e34292.
January 31, 2018
Apply to the most prestigious workshop in Molecular Evolution at the MBL in Woods Hole, July 19 – July 28! I will be part of the faculty crew this year to talk about multi-gene phylogenetics for deep divergence.
MBL’s Workshop on Molecular Evolution is the most prestigious workshop serving the field of evolutionary studies. Founded in 1988, it is the longest-running workshop if its kind, and it has earned worldwide recognition for its rich and intensive learning experience. Students work closely with internationally-recognized scientists, receiving (i) high-level instruction in the principles of molecular evolution and evolutionary genomics, (ii) advanced training in statistical methods best suited to modern datasets, and (iii) hands-on experience with the latest software tools (often from the authors of the programs they are using). The material is delivered via lectures, discussions, and bioinformatic exercises motivated by contemporary topics in molecular evolution. A hallmark of this workshop is the direct interaction between students and field-leading scientists. The workshop serves graduate students, postdocs, and established faculty from around the world seeking to apply the principles of molecular evolution to questions of anthropology, conservation genetics, development, behavior, physiology, and ecology. The workshop also welcomes participants from federal agencies and science journalists. A priority of this workshop is to foster an environment where students can learn from each other as well from the course faculty.
Faculty members include:
Cécile Ané, University of Wisconsin-Madison
Peter Beerli, Florida State University
Joseph Bielawski, Dalhousie University
Belinda Chang, University of Toronto
Casey Dunn, Yale University
Scott Edwards, Harvard University
Laura Eme, Uppsala University
Tracy Heath, Iowa State University
Mark Holder, University of Kansas
John Huelsenbeck, University of California-Berkeley
Lacey Knowles, University of Michigan
Laura Kubatko, Ohio State University
Michael Landis, Yale University
Paul Lewis, University of Connecticut
Emily Jane McTavish, University of California-Merced
Bui Quang Minh, Centre for Integrative Bioinformatics, Vienna
David Swofford, Duke University
Rachel Williams, Duke University
Anne Yoder, Duke University
27 October 2017
Submit and take part in our symposium at the II Joint Congress on Evolutionary Biology.
– New approaches to phylogenomics –
Organizers: Laura Eme, Carolin Kosiol, Vincent Daubin, Nicola De Maio
Genomes maintain a chronicle of their own history, from short scale evolution (e.g. cancer and within-host viral evolution) to the last common ancestor of all extant life. For some organisms, such as most microbial lineages, it is practically the sole source of evolutionary information available. For others, the interdependence between genome evolutionary patterns and life history traits (phenotype, ecology, epigenetic…) documented using other techniques remains largely obscure. Recent methodological developments in comparative genomics allow for gene tree / species tree reconciliation and the identification of horizontal gene transfer, for estimating the age, polymorphism and gene repertoire of ancestral populations, for tracing back the history of migrations, population splits and admixtures, and for reconstructing mutational and selective pressures. Understanding the link of these patterns to life history traits, population dynamics and evolutionary success remains a major question and methodological challenge. This symposium is dedicated to new methodological developments in integrating genomic data with other sources of information, from fossils, to phenotypic, epidemiological, epigenetic and geographical data.
Invited speaker: Andrew J. Roger “Realism in phylogenetic models is essential for reconstructing early eukaryote evolution”.
Note the possibility to apply for financial aid for female attendees!
11 September 2017
Blastocystis is the most prevalent eukaryotic microbe colonizing the human gut, infecting approximately 1 billion individuals worldwide. In this collaborative work, we have sequenced and analyzed the genome of Blastocystis subtype 1, and compared to previously published sequences for ST4 and ST7. Overall, this study provides an important window into the biology of Blastocystis, showcasing significant differences between STs that can guide future experimental investigations into differences in their virulence and clarifying the roles of these organisms in gut health and disease.
Extreme genome diversity in the hyper-prevalent parasitic eukaryote Blastocystis. PLos biology 15 (9), e2003769.
08 August 2017 – Our outreach piece on eukaryogenesis is out in Microbiology Today.
The symbiosis that changed the world. Laura Eme & Thijs J.G. Ettema
01 August 2017 – Currently at the International Congress of Protistology (Prague, Czech republic). Will be part of the ‘Origin of eukaryotes’ session, and will talk about the archaeal roots of eukaryotes and new Asgard data, Thursday @14.00 in the Labe hall.
24 July 2017 – Currently at the Gordon Research Conference Archaea.
I will give a talk on Monday @8:50 pm about the archaeal origins of eukaryotes and eukaryogenesis.
In addition, come and discuss my poster (#58) entitled ‘Asgard archaea are the closest archaeal relatives of eukaryotes’.
13 Feb 2017 – Early career researcher interested in symbiosis? Apply for a funded place to attend our workshop “Symbiosis in the microbial world: from ecology to genome evolution“! It will be held in West Sussex, UK from November 5th to 9th, 2017.
This informal workshops will gather 25 world-wide experts and 10 early-career participants and will take place in the beautiful Wiston House in Wilton Park (www.wistonhouse.co.uk).






